Difference between revisions of "Proteomics and Proteogenomics"
Debra Tabron (talk | contribs) |
m (Protected "Proteomics and Proteogenomics" ([Edit=Allow only administrators] (indefinite) [Move=Allow only administrators] (indefinite))) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
Proteomics is the analysis of proteins present in a sample. Proteogenomics is the combined use of proteomics with genomics and transcriptomics to support protein identifications and analyses. As tools, proteomics and proteogenomics allow researchers and practitioners to understand the functional gene products and relevant microbial metabolisms in a system, which in turn can lead to informed decision-making in remediation situations. | Proteomics is the analysis of proteins present in a sample. Proteogenomics is the combined use of proteomics with genomics and transcriptomics to support protein identifications and analyses. As tools, proteomics and proteogenomics allow researchers and practitioners to understand the functional gene products and relevant microbial metabolisms in a system, which in turn can lead to informed decision-making in remediation situations. | ||
<div style="float:right;margin:0 0 2em 2em;">__TOC__</div> | <div style="float:right;margin:0 0 2em 2em;">__TOC__</div> | ||
| + | |||
'''Related Article(s):''' | '''Related Article(s):''' | ||
Latest revision as of 13:54, 7 January 2022
Proteomics is the analysis of proteins present in a sample. Proteogenomics is the combined use of proteomics with genomics and transcriptomics to support protein identifications and analyses. As tools, proteomics and proteogenomics allow researchers and practitioners to understand the functional gene products and relevant microbial metabolisms in a system, which in turn can lead to informed decision-making in remediation situations.
Related Article(s):
Contributor(s): Dr. Kate Kucharzyk, Dr. Morgan Evans, Dr. Robert Murdoch, Dr. Fadime Murdoch, and Larry Mullins
Key Resources(s):
Introduction
Proteomics is the comprehensive analysis of the proteins produced by single organisms or by a microbial community (e.g., “meta”-proteomics). In this regard, proteomics represents the identification of functional gene products, providing information and insight into structural proteins and the molecular machinery produced and utilized by organisms to sustain the metabolic processes. While proteomics data can be analyzed in a de novo manner by comparing to global protein sequence databases, from a systems biology perspective, the ideal starting point for all considerations and the key enabling information is the (meta)genome of the sample under study.
Proteogenomics combines proteomics with (meta)genomics and/or transcriptomics to better analyze and identify proteins[2]. For environmental studies, proteomics can be applied to soil, groundwater, sediment, or other environmental samples. Proteomics allows for functional characterization of a sample, enabling investigators to infer what relevant metabolical pathways may be active in a system (e.g., hydrocarbon degradation, reductive dechlorination). The large-scale characterization of any given proteome is accomplished by comparing measured peptide spectra with predicted protein or peptide data derived from (meta)genomic information. Thus, it is vital to have complete (meta)genome sequence information for the system being studied [3].This approach coined the term proteogenomics which describes the strong linkage between genomics and proteomics. As implied, the quality of the proteomic measurements is inextricably linked to the quality of the genomic or metagenomic sequence data. Proteogenomics is an inherently more uncertain technique when compared to nucleic acid sequencing or qPCR technologies, yet it provides unparalleled global insights into biological structure and function.
DNA-based methods such as shotgun metagenomics and 16S rRNA amplicon sequencing provide important information and guidance for potential function of microbial communities. Like shotgun proteomics, both methods also provide relative abundances of many features. On the other hand, quantitative real time PCR (qPCR) provides absolute quantities of few features with greater sensitivity (at least 1 order of magnitude) than metagenomics approaches[4]. As with all nucleic-acid-based (eg. DNA or RNA) methods, qPCR only informs potential activity, not actual activity. By detecting gene expression rather than simply genes, RNA-based methods such as metatranscriptomics provide insight into which genes are active[5], but at the cost of additional challenges posed by increased difficulty with RNA isolation and instability. Proteomics provides the actual catalytic activity by detection and quantification of proteins of interest.
Advantages
The two main advantages of the application of metaproteomics are:
- the ability to directly measure microbial enzymes (not just potential for enzyme synthesis), and
- the ability to generate detailed information on hundreds of microorganisms and proteins in one assay.
The process of obtaining the information does not require culturing of microorganisms or performance of any molecular assays. To measure features that are directly correlated to microbial activity, metaproteomics can be used to identify and relatively quantify proteins[6][7][8]. These proteins provide important information about community activities, such as which microbial organisms are most active and what proteins are present (including proteins catalyzing reactions involved in bioremediation)[1][9].
Limitations
Limitations associated with this technology are related to composition of the proteome to be analyzed, mainly concerning protein expression levels and limitations of the analytical equipment. Limitations are summarized as follows:
- Sample preparation - the vast diversity of protein molecular sizes, charge states, conformational states, post-translational modifications etc., make it unfeasible to use a single sample preparation protocol that captures the entire proteome for a given microbial community. Thus, use of a protocol that allows isolation of protein content from a biological sample and eliminates non-specific contaminants (e.g., keratins, fatty acids, plastic polymers, nucleic acids and salt clusters) should be developed and tested prior to sample analysis[10][11]
- Proteins are not expressed in equal amounts and there may be large differences in protein levels in proteomes in samples collected from the same site. A proteomic analysis must employ proper analytical techniques for the detection of all proteins or proteins of interest. In a small sample volume that is usually used in a proteomic analysis, a large percentage of the expressed proteins occur at low abundance levels and cannot be readily detected in the analysis due to high-abundance proteins effectively monopolizing the “sampling effort”. These low abundant proteins may be of particular interest in environmental samples because proteins associated with contaminant degradation are often a very small fraction of the total expressed proteins. The practical protein detection limit for Liquid Chromatography-Tandem Mass Spectrometry-Time-of-Flight (LC-MS/MS-TOF) analysis lies in the femtomol (10-15 [fmol]) range. However, due to losses during protein extraction and sample clean up and dilution, the sufficient protein concentration for detection is more realistically in the low picomol (10-12 [pmol]) to high fmol range. This limitation can be addressed by collecting a greater volume of groundwater for analysis; however, this may not be possible at all sampling locations.
- Detection of proteins at low concentrations (low abundance) may be limited by other proteins present in high concentrations (high abundance). The successful search for low abundant proteins may be mitigated by use of chromatography for separation of high-abundance proteins and precipitation for elution of proteins of interest prior to analytical detection. However, complete removal of highly abundant proteins may not be recommended because they may trap the low-abundance proteins along with their associated fragments and peptides, which will be lost and not detected. An alternative approach relies on 2-dimensional chromatography coupled with tandem mass spectrometry.
- Success in the identification of proteins may vary with the sensitivity of the mass spectrometer. Proper analytic equipment can be costly. Of the most sensitive mass spectrometers, electrospray ionization and laser desorption ionization-based instruments can detect peptides with low detection limits.
- Not all biological variation can be accounted for. Despite great improvements in the costs of genomic sequencing and gene prediction, there remain some aspects of biology that cannot be accurately or consistently predicted. For example, the presence and variety of post-translational modifications remains problematic and, if present and not accounted for during analysis, such modified proteins will not be efficiently detected.
- Selection of the appropriate analytical method determines the success of the study. Depending on the study goals and context, some methodological adjustments should be considered, including pre-enrichment of key low-abundance proteins, adjustment of protein extraction methods or tuning of the analytical equipment. A single methodological approach is not suitable for all purposes.
Assessing Changes in Microbial Community Composition and Dynamics – Common Applications
Environmental metaproteomics is used in applied research areas such as:
- Bioenergy – characterization of feedstock conversion into energy e.g., cellulose or lignin degradation to biofuels[12]
- Human health – characterization of microbial involvement in impact/control of disease versus health in human bodies[13][14]
- Bioremediation – characterization of degradation of contaminants in sediments, soils, and groundwater by microorganisms[15][16][17][18][19][20]
- Carbon cycling – characterization of the role of microorganisms in carbon flow in an ecosystem.
- Agricultural metabolism – characterization of microbial interactions with plants[21]
Targeted vs Shotgun Proteomics
Proteomics techniques can be broadly classified into two categories:
- Untargeted or shotgun proteomics, aimed at comprehensively identifying and characterizing relative abundances of the totality of proteins in a sample.
- Targeted proteomics, focused on identifying and absolutely quantifying one protein.
Shotgun proteomics refers to digestion of the total proteome and subjecting all resulting peptides to separation, mass-spectroscopy, and identification based on a reference protein database (ideally derived from in silico translation of the (meta)genome). Notably, shotgun proteomics generates only an approximated relative abundance for identified proteins. On the other hand, targeted proteomics allows for absolute quantification of a single protein within a complex sample, which in turn allows for analysis of any potential correlation to a degradation rate. Prediction of degradation rates based on enzyme concentration is a crucial step towards better understanding of the molecular events underlying metabolic processes. By measuring key biomarkers, proteomic studies present an opportunity to gain profound insight into ecosystem health, degradation of recalcitrant compounds, and bioremediation. Quantitative proteomics can also guide regulatory agencies to make better site management decisions, thereby minimizing radiation costs and chemically induced adverse effects.
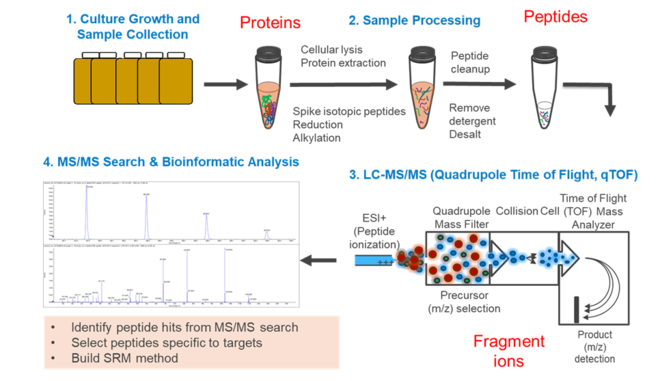
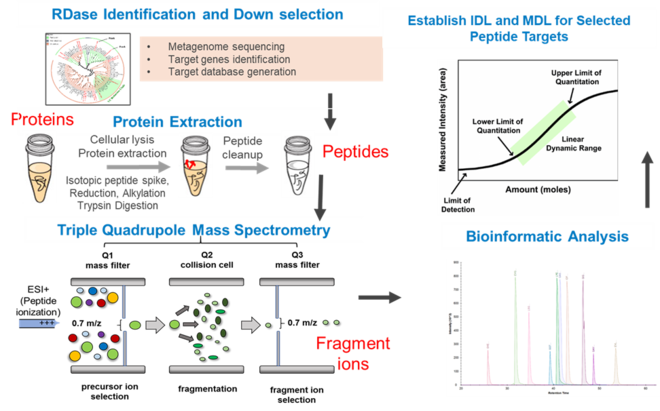
Targeted Proteomics targets peptides of a specific protein in a complex mixture of other peptides and determines their presence (if they are above the detection limit) and quantity in one sample or across multiple samples (Figure 1)[22]. This analysis usually utilizes a triple quadrupole mass spectrometer (QqQ-MS), an instrument which has traditionally been used to quantify small molecules. Only recently has it been utilized for peptides. Parallel Reaction Monitoring (PRM) and data independent analysis are alternative options that can be far cheaper than developing a method for a QqQ-MS. Each targeted proteomics assay must be carefully developed. The assay specifically “targets” peptides enzymatically digested from the target protein, necessitating careful selection of peptides and method. Development of a new assay also requires preliminary analyses to ensure the method is robust. Once a method is developed and verified, it can be applied to an unlimited number of samples.
Bottom-Up Proteomics
Bottom-up proteomics (Figure 2) begins by analyzing of mass spectrum fragmentation patterns of peptides, which are generated after proteolytic digestion of proteins[22]. Spectra are identified by comparison to a database of reference proteins which are digested in silico. The creation or selection of this database is one of the most crucial steps in the analysis; ideally, the database consists of proteins encoded by the genome or metagenome of the organisms present in the sample under study. While the rapidly decreasing cost of DNA sequencing and emergence of standard assembly and annotation pipelines make obtaining (meta)genomes increasingly cost-effective, it remains an option to use global reference protein databases, such as UniProt (see unipept.ugent.be). However, making peptide-spectrum matches (a.k.a. PSMs) is a statistically uncertain process, as the alignment of mass fragmentation pattern of the peptide to the database is seldom perfect. Several algorithms have been developed to tackle this problem (Comet, X! Tandem, MyriMatch, OMSSA, Tide, etc.). PSM matching must carefully consider factors such as what enzyme was used to digest the proteome and how specific and complete digestion was. Additionally, peptide modifications, chemical modification of amino acids which lead to differences in mass and fragmentation pattern, are a wide-spread complication that must be accounted for. Some are intentional, for example carbiodomethylation of cysteine residues, which is applied to protect sulfur residues from becoming oxidized. Other modifications occur unintentionally during sample preparation, such as oxidation of methionine residues. PSMs are inherently statistically uncertain. A given shotgun proteome may involve hundreds of thousands of PSMs, which leads to the danger of false discovery. Traditionally, a formal false discovery rate (FDR) is applied; this is an adjustment of the threshold for statistical significance based on the number of tests performed, i.e. the more PSMs, the more stringent the testing must be. The risk is greatly exacerbated by using reference protein databases that do not reflect the sample. Use of the (meta)genome correlating to the sample under study makes this danger of false identification much less risky when compared to traditional approaches. It is generally acknowledged that rigid adherence to FDR thresholds is a serious impediment to thorough PSM assignment[23] but is generally advisable when analyzing a proteome without any pre-existing knowledge on sample composition. Following PSM assignment, whether by use of a global reference protein database with application of FDR or by comparison to a sample-specific reference (meta)genome, peptides are matched to proteins. This step acts as a second statistical filtering step and can employ several criteria such as whether the peptide identified is unique in the database, how many peptides match a given protein, and what score was assigned to the PSMs by the PSM algorithm. A metaproteome protein identification might, for example, require three or six matching PSMs.
Software for making PSMs and protein identifications can be obtained as individual packages, but several convenient open-source packages are available, some of which include several PSM algorithms (SearchGUI) and even pipelines for making further sample comparisons (PatternLab for Proteomics) or functional interpretations (MetaProteomeAnalyzer). Many of these features are also available in commercial software packages, such as Progenesis QI (Waters) and ProteinPilot (SCIEX). Global bottom-up shotgun proteomics data analysis remains an actively evolving discipline.
Selecting Sample Locations
Below are a few guidelines for selecting sampling locations to aid in drawing conclusions from metaproteomics data.
- Background: Samples from non-impacted background area can be compared with results from impacted areas to examine the impact of contamination on composition of microbial proteomes that reflect the ongoing metabolical processes.
- Baseline: These samples are collected and analyzed prior to treatment as a baseline for evaluating changes in the microbial metabolism in response to the remediation.
- Plume: These samples are collected from distinct zones within the source area or contaminant plume to reflect variations in contaminant concentrations, geochemical conditions, and other site-specific criteria.
Sample Collection, Preservation, and Shipping
Sampling procedures for proteomics analyses are straightforward and readily integrated into existing monitoring programs. Almost any type of sample matrix (soil, sediment, groundwater) or filters (on-site filtration) can be analyzed. All samples should be shipped to the laboratory on ice or dry ice (-20 °C) using an overnight carrier to minimize the potential for changes in the microbial community between collection and analysis and keep integrity of proteins. Groundwater samples (typically 1 L) can be shipped directly to the laboratory or filtered in the field. For on-site filtration, groundwater is pumped through a Sterivex® or Bio-Flo® filter using standard low flow sampling techniques. The groundwater may then be discarded appropriately. As with other sample types, filters should be shipped on ice (4 °C) using an overnight carrier.
References
- ^ 1.0 1.1 Arsène-Ploetze, F., Bertin, P.N., and Carapito, C., 2015. Proteomic tools to decipher microbial community structure and functioning. Environmental Science and Pollution Research, 22, pp. 13599-13612. DOI: 10.1007/s11356-014-3898-0 Article pdf
- ^ Helbling, D.E., Ackermann, M., Fenner, K., Kohler, H.P., and Johnson, D.R., 2012. The activity level of a microbial community function can be predicted from its metatranscriptome. The ISME Journal, 6(4), pp. 902-904. doi: 10.1038/ismej.2011.158 Article pdf
- ^ Ansong C., Purvine, S.O., Adkins, J.N., Lipton, M.S., and Smith, R.D., 2008. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Briefings in Functional Genomics, 7(1), pp. 50-62. doi: 10.1093/bfgp/eln010 Article pdf
- ^ Clark K., Taggart, D.M., Baldwin, B.R., Ritalahti, K.M., Murdoch, R.W., Hatt, J.K., and Löffler, F.E., 2018. Normalized Quantitative PCR Measurements as Predictors for Ethene Formation at Sites Impacted with Chlorinated Ethenes. Environmental Science & Technology, 52(22), pp. 13410-13420. doi: 10.1021/acs.est.8b04373
- ^ Czaplicki, L.M. and Gunsch, C.K., 2016. Reflection on Molecular Approaches Influencing State-of-the-Art Bioremediation Design: Culturing to Microbial Community Fingerprinting to Omics. Journal of Environmental Engineering, 142(10), pp. 1-13. doi: 10.1061/(ASCE)EE.1943-7870.0001141
- ^ Hettich, R.L., Pan, C., Chourey, K., and Giannone, R.J., 2013. Metaproteomics: Harnessing the Power of High Performance Mass Spectrometry to Identify the Suite of Proteins That Control Metabolic Activities in Microbial Communities. Analytical Chemistry, 85(9), pp. 4203-4214. doi: 10.1021/ac303053e Article pdf
- ^ Keller, M. and Hettich, R.L., 2009. Environmental Proteomics: a Paradigm Shift in Characterizing Microbial Activities at the Molecular Level. Microbiology and Molecular Biology Reviews, 73(1), pp. 62-70. doi: 10.1128/MMBR.00028-08 Article pdf
- ^ Schneider, T. and Riedel, K., 2010. Environmental proteomics: Analysis of structure and function of microbial communities. Proteomics, 10(4), pp. 785-98. doi: 10.1002/pmic.200900450
- ^ Johnson, D.R., Helbling, D.E., Men, Y., and Fenner, K., 2015. Can meta-omics help to establish causality between contaminant biotransformations and genes or gene products? Environmental Science: Water Research & Technology, 1, pp. 272-278. doi: 10.1039/C5EW00016E Article pdf
- ^ Chourey, K., Jansson, J., VerBerkmoes, N., Shah, M., Chavarria, K.L., Tom, L.M., Brodie, E.L., and Hettich, R.L., 2010. Direct Cellular Lysis/Protein Extraction Protocol for Soil Metaproteomics. Journal of Proteome Research, 9(12), pp. 6615-6622. doi: 10.1021/pr100787q
- ^ Qian, C. and Hettich, R.L., 2017. Optimized Extraction Method To Remove Humic Acid Interferences from Soil Samples Prior to Microbial Proteome Measurements. Journal of Proteome Research, 16(7), pp. 2537-2546. doi: 10.1021/acs.jproteome.7b00103
- ^ Ndimba, B.K., Ndimba, R.J., Johnson, T.S., Waditee-Sirisattha, R., Baba, M., Sirisattha, S., Shiraiwa, Y., Agrawal, G.K., and Rakwal, R, 2013. Biofuels as a sustainable energy source: an update of the applications of proteomics in bioenergy crops and algae Journal of Proteomics, 20(93), pp. 234-244. doi: 10.1016/j.jprot.2013.05.041
- ^ Brooks, B., Mueller, R.S., Young, J.C., Morowitz, M.J., Hettich, R.L., and Banfield, J.F., 2015. Strain-resolved microbial community proteomics reveals simultaneous aerobic and anaerobic function during gastrointestinal tract colonization of a preterm infant. Frontiers in Microbiology, 1(6) pp. 654. doi: 10.3389/fmicb.2015.00654 Article pdf
- ^ Carr, S.A., Abbatiello, S.E., Ackermann, B.L., Borchers, C., Domon, B., Deutsch, E.W.,, Grant, R.P., Hoofnagle, A.N., Hüttenhain, R., Koomen, J.M., Liebler, D.C., Liu, T., MacLean, B., Mani, D.R., Mansfield, E., Neubert, H., Paulovich, A.G., Reiter, L., Vitek, O., Aebersold, R., Anderson, L., Bethem, R., Blonder, J., Boja, E., Botelho, J., Boyne, M., Bradshaw, R.A., Burlingame, A.L., Chan, D., Keshishian, H., Kuhn, E., Kinsinger, C., Lee, J.S., Lee, S.W., Moritz, R., Oses-Prieto, J., Rifai, N., Ritchie, J., Rodriguez, H., Srinivas, P.R., Townsend, R.R., Van Eyk, J., Whiteley, G., Wiita, A., and Weintraub, S., 2014. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Molecular and Cellular Proteomics, 13(3), pp. 907-917. doi: 10.1074/mcp.M113.036095 Article pdf
- ^ Bansal, R., Deobald, L.A., Crawford, R.L., Paszczynski, A.J., 2009. Proteomic detection of proteins involved in perchlorate and chlorate metabolism. Biodegradation, 20(5), pp.603-620. doi: 10.1007/s10532-009-9248-0
- ^ Fuller, M. E., van Groos, P. G. K., Jarrett, M., Kucharzyk, K. H., Minard-Smith, A., Heraty, L. J., and Sturchio, N. C., 2020. Application of a multiple lines of evidence approach to document natural attenuation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) in groundwater. Chemosphere. 250, pp. 126210. doi: 10.1016/j.chemosphere.2020.126210
- ^ Kucharzyk, K. H., Meisel, J. E., Kara-Murdoch, F., Murdoch, R. W., Higgins, S. A., Vainberg, S., and Löffler, F. E., 2020. Metagenome-guided proteomic quantification of reductive dehalogenases in the Dehalococcoides mccartyi-containing consortium SDC-9. Journal of Proteome Research, 9(4), pp. 1812-1823. doi: 10.1021/acs.jproteome.0c00072
- ^ Kucharzyk, K.H., Rectanus, H.V., Bartling, C., Chang, P., Rosansky. S., Neil, K., and Chaudhry, T., 2018. Assessment of Post Remediation Performance of a Biobarrier Oxygen Injection System at a Methyl Tert-Butyl Ether (MTBE)-Contaminated Site, Marine Corps Base Camp Pendleton San Diego, California. Environmental Security Technology Certification Program (ESTCP), Alexandria, VA. ER-201588. Report pdf
- ^ Michalsen, M. M., Kucharzyk, K. H., Bartling, C., Meisel, J. E., Hatzinger, P., Wilson, J., Istok, J., and Loffler, F, 2020. Validation of advanced molecular biological tools to monitor chlorinated solvent bioremediation and estimate cVOC degradation rates. Environmental Security Technology Certification Program, Alexandria, VA. ER-201726. Report pdf
- ^ Michalsen, M. M., King, A. S., Istok, J. D., Crocker, F. H., Fuller, M. E., Kucharzyk, K. H., and Gander, M. J., 2020. Spatially distinct redox conditions and degradation rates following field-scale bioaugmentation for RDX-contaminated groundwater remediation. Journal of Hazardous Materials, 387, 121529. doi: 10.1016/j.jhazmat.2019.121529
- ^ Tan, B.C, Lim, Y.S., and Lau, S.E., 2017. Proteomics in commercial crops: An overview. Journal of Proteomics, 169, pp. 176-188. doi:10.1016/j.jprot.2017.05.018
- ^ 22.0 22.1 22.2 Zhang, Y., Fonslow, B.R, Shan, B., Baek, M.C., and Yates, J.R. III., 2013. Protein analysis by shotgun/bottom-up proteomics. Chemical Reviews, 113(4), pp 2343-94. doi:10.1021/cr3003533
- ^ Heyer, R., Schallert, K., Zoun, R., Becher, B., Saake, G., and Benndorf, D., 2017. Challenges and perspectives of metaproteomic data analysis. Journal of Biotechnology, 261, pp. 24–36. doi: 10.1016/j.jbiotec.2017.06.1201 Article pdf
See Also
- Strategies for Metagenomic-Guided Whole-Community Proteomics of Complex Microbial Environments
- Options and considerations when selecting a quantitative proteomics strategy
- Environmental OMICS: Current Status and Future Directions
- An Alignment-Free “Metapeptide” Strategy for Metaproteomic Characterization of Microbiome Samples Using Shotgun Metagenomic Sequencing
- Environmental proteomics: A long march in the pedosphere
- Proteome profile and proteogenomics of the organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1 grown on hexachlorobenzene as electron acceptor
- Unipept 4.0: Functional Analysis of Metaproteome Data
- Guidelines for reporting the use of mass spectrometry in proteomics
- The minimum information about a proteomics experiment (MIAPE)
- Next-Generation Proteomics: Toward Customized Biomarkers for Environmental Biomonitoring
- A decade of metaproteomics: Where we stand and what the future holds